About

Bernardo Almeida
AI Senior Research Scientist at InstaDeep
My broader research interests include Deep Learning, Genomics and Personalized Medicine. I previously completed my PhD at the Research Institute of Molecular Pathology (IMP) in Vienna, working with Alex Stark on using deep learning and massively parallel reporter assays to decode the regulatory grammar of regulatory DNA sequences.
News
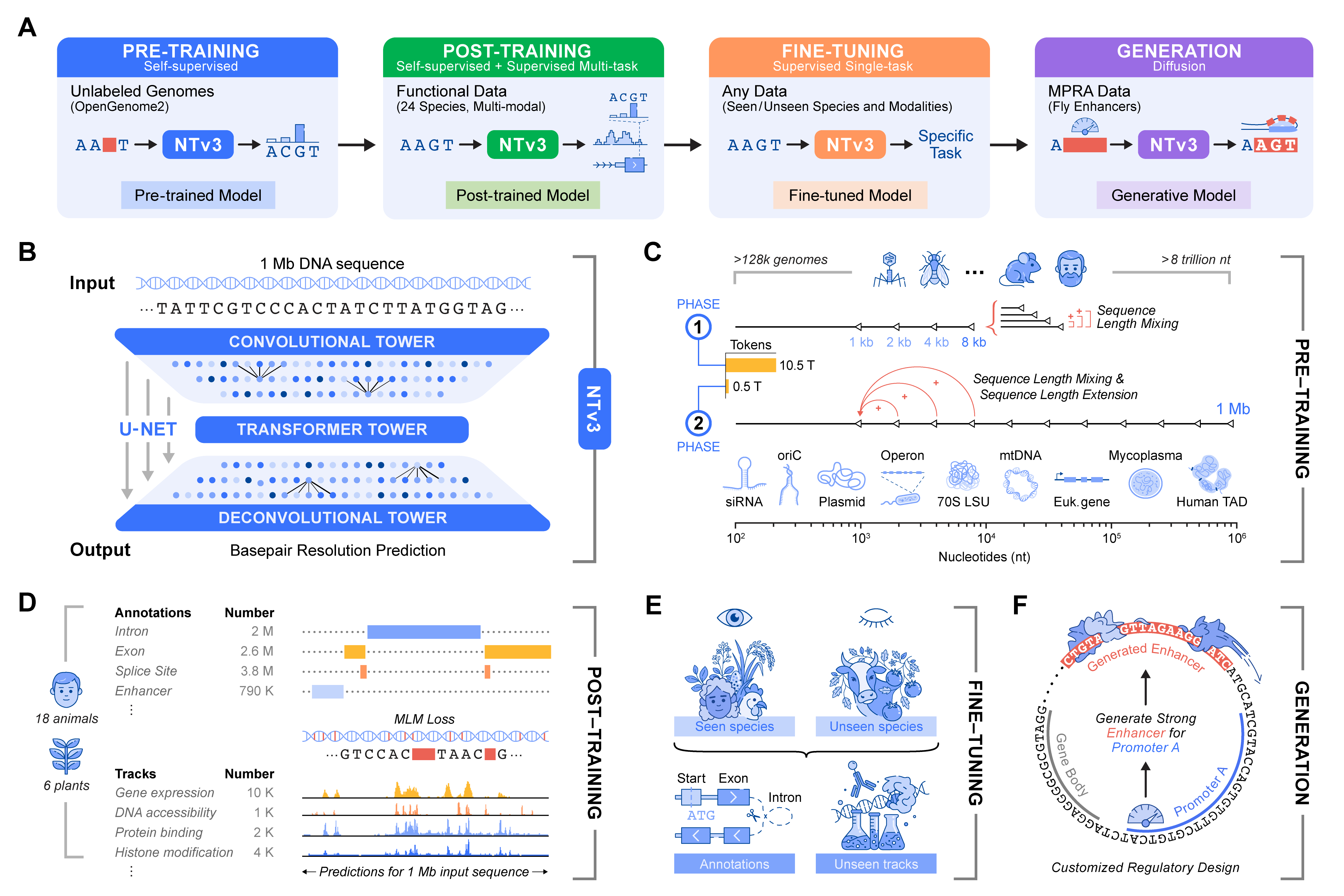
We are excited to introduce Nucleotide Transformer v3 (NTv3) - InstaDeep's new foundation model for long-range multi-species genomics 🎉
"Annotating the genome at single-nucleotide resolution" accepted to Nature Methods
We just open-sourced the first large-scale dataset designed to train conversational agents on biological sequences 🧬

Our paper "A multimodal conversational agent for DNA, RNA and protein tasks" was featured on the cover of Nature Machine Intelligence!
The Nucleotide Transformer paper is now published in Nature Methods
Our work on the "Nucleotide Transformer: building and evaluating robust foundation models for genomics" was presented at CSHL Systems Biology (March 2024) and MLCB (September 2024) conferences

Very happy to receive 2024 Denise P. Barlow Award for best PhD thesis on biological mechanisms in Vienna, Austria (May 2024)
For more information see here


It was great to participate in the podcast Futuro do Futuro (in portuguese) and discuss my work and the future of AI in biology (December 2023)
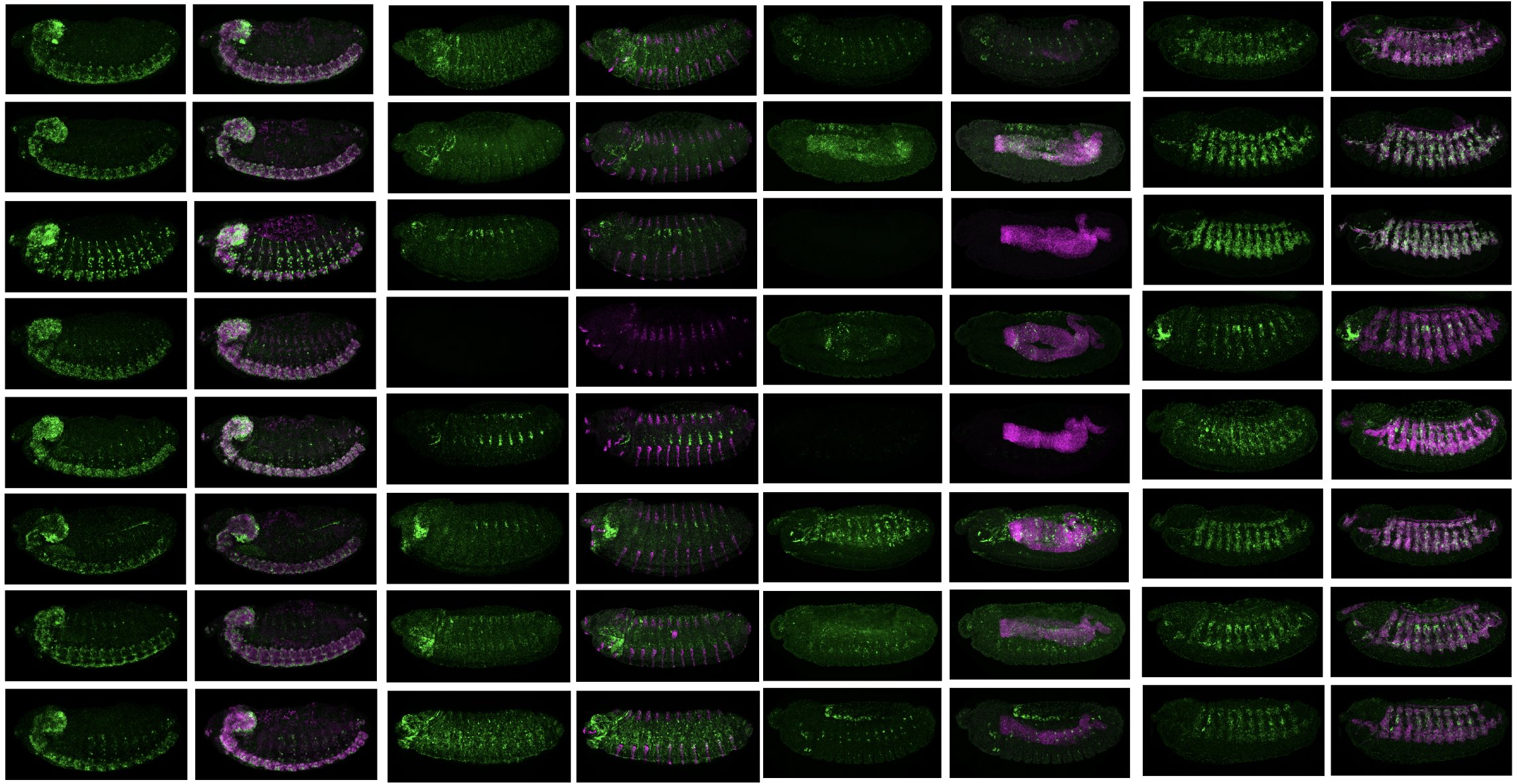
My final PhD paper on designing synthetic enhancers for selected tissues in vivo using AI is out in Nature (December 2023)
I am very happy to have received one of the 2023 Vienna BioCenter PhD Awards! Shared with great colleagues. It's the best way to close the PhD at the IMP, in Vienna (November 2023)


Very happy to give a OLISSIPO Workshop on Decoding the genome with deep learning, including a hands-on session (April 2023)
Check the Kipoi Seminar where I presented my work on Decoding the cis-regulatory information of enhancer sequences (April 2023)


Very honoured to receive The Life Science Research Award Austria 2022 from the Austrian Society for Molecular Biosciences and Biotechnology (ÖGMBT) in the category "Basic Science" (September 2022)
For more information see the official press release
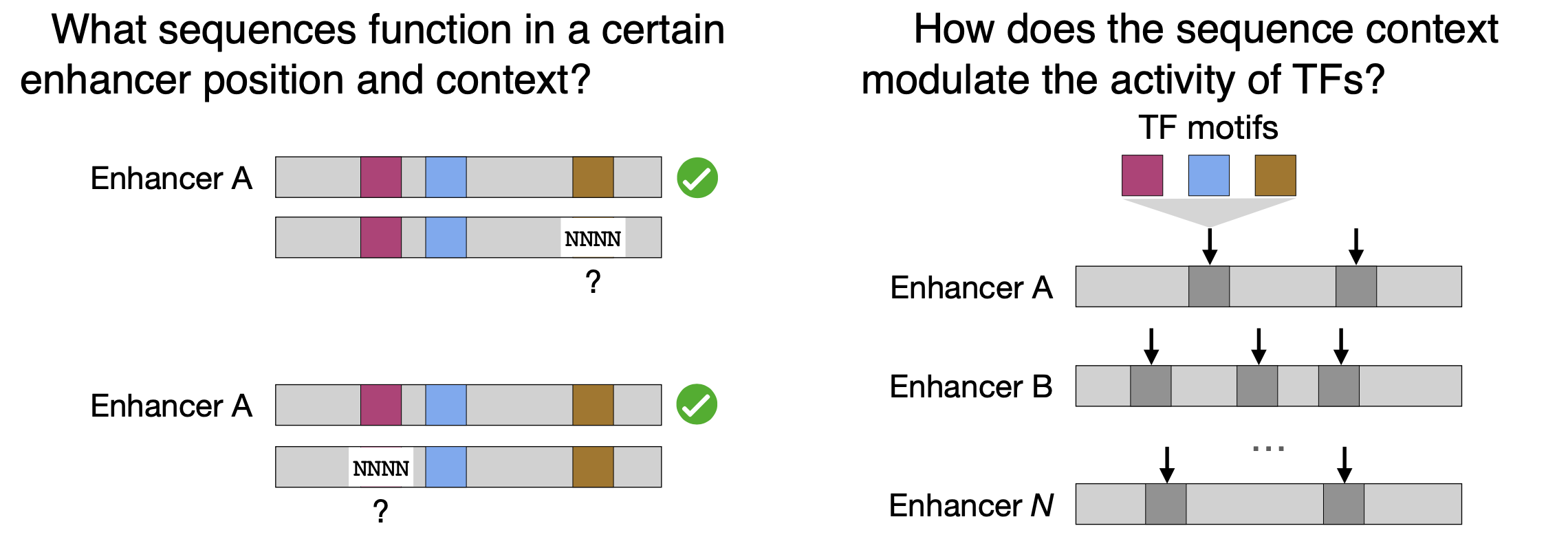
Presented a poster at the 15th EMBL Conference: Transcription and Chromatin about our new preprint on the flexibility of enhancer sequences and motif syntax (August 2022)
Check out the poster here
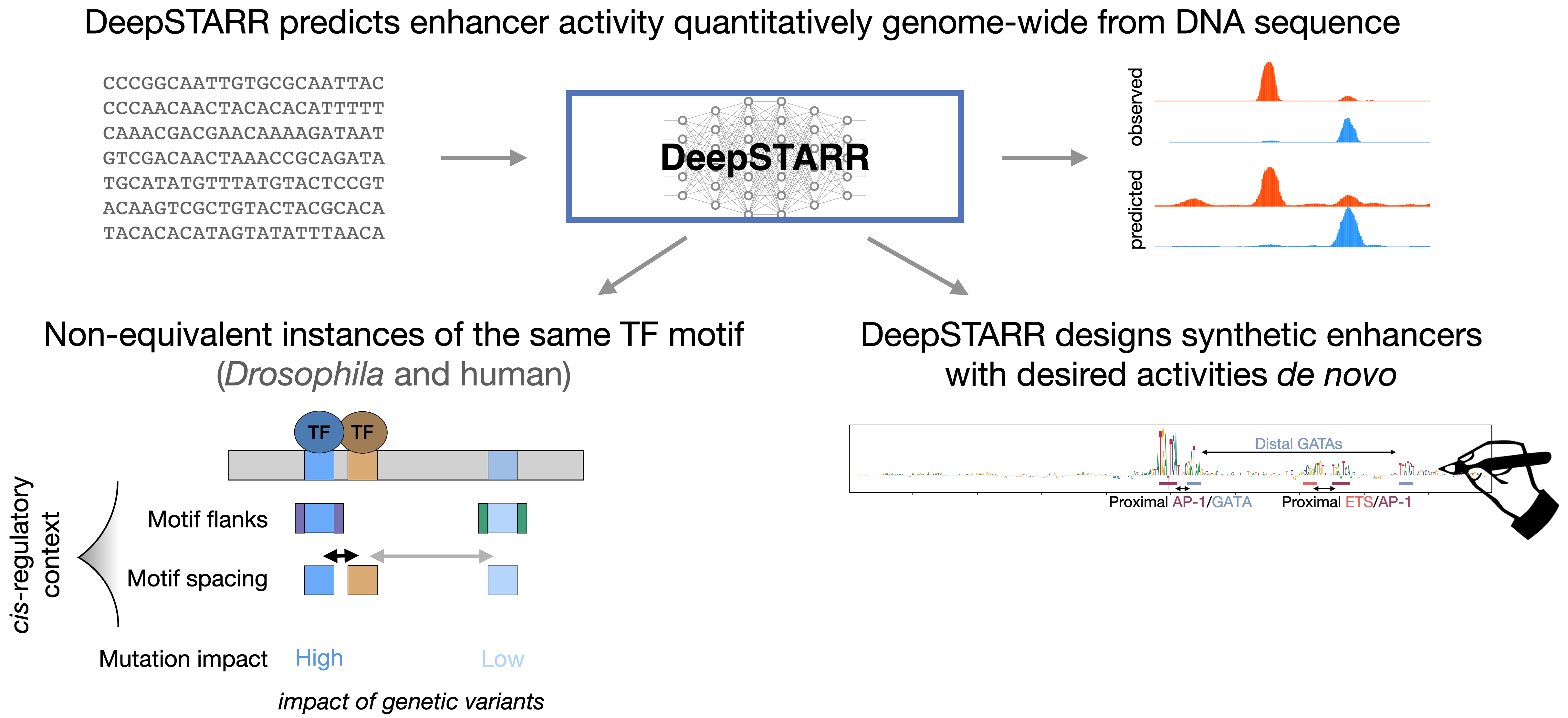
Very happy to see our DeepSTARR paper highlighted in Nature Methods: "Predicting and designing enhancers" (July 2022)
Check the ISCBacademy Webinar where I presented my recent work DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers (December 2021)