Tutorials of deep learning models
DeepSTARR: a multi-task deep learning model (CNN) to predict enhancer activities genome-wide.

DeepSTARR is a multi-task CNN built to quantitatively predict the activities of developmental and housekeeping enhancers from DNA sequence in Drosophila melanogaster S2 cells. Enhancer activities were assessed genome-wide using the transcriptional reporter assay STARR-seq.
This tutorial will show you how to (1) train DeepSTARR, (2) compute nucleotide contribution scores, and (3) discover predictive motifs with TF-MoDISco.
Enformer: a transformer-based deep learning model (based on self-attention) with the ability to predict chromatin data and gene expression from DNA sequences.


The model was trained to predict thousands of epigenetic and transcriptional dataset from both human and mouse in a multitask setting across long DNA sequences.
This tutorial will show you how to (1) use the pre-trained Enformer model to make predictions of different chromatin states and gene expression for a given locus, (2) compute nucleotide contribution scores for those predictions, and (3) predict the effect of a genetic variant on those profiles.
This tutorial is based on the enformer-usage colab.
Other tutorials
Self-Attention: this tutorial will show you how Self-Attention works and use it to solve a simple task: given an array of 10 integers (0-9), predict wether there are more 2s than 4s.
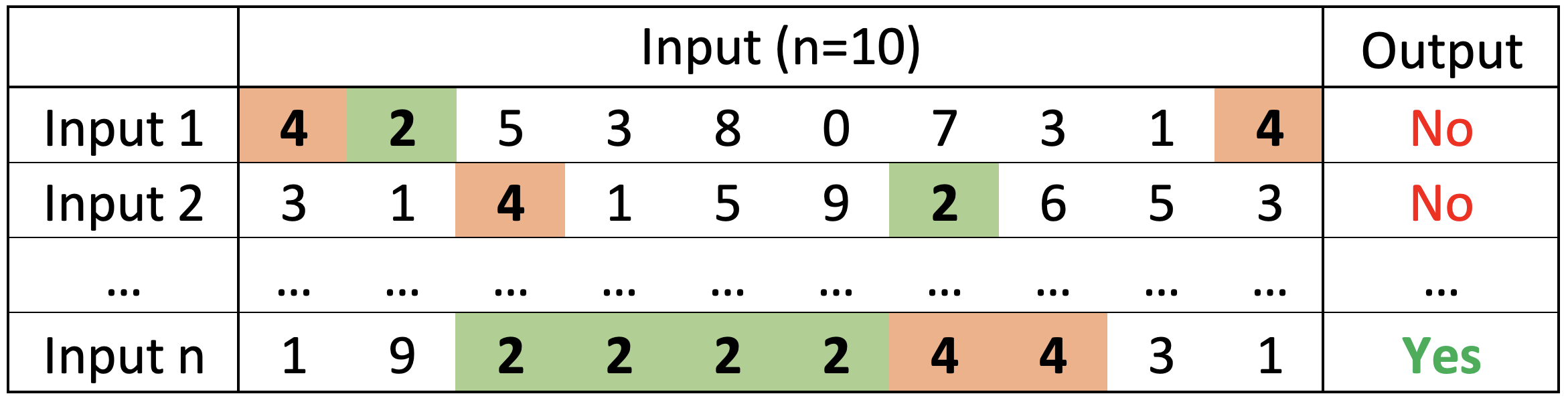
This tutorial will show you how to (1) code the different Self-Attention calculations, (2) implement a Self-Attention layer from scratch or using the built-in Keras Attention layer, and (3) train simple models and inspect the Attention weights and values learned during training.