Publications
Selected
21. B.P. de Almeida, C. Schaub, M. Pagani, S. Secchia, E.E.M. Furlong, A. Stark. Targeted design of synthetic enhancers for selected tissues in the Drosophila embryo. Nature 2024
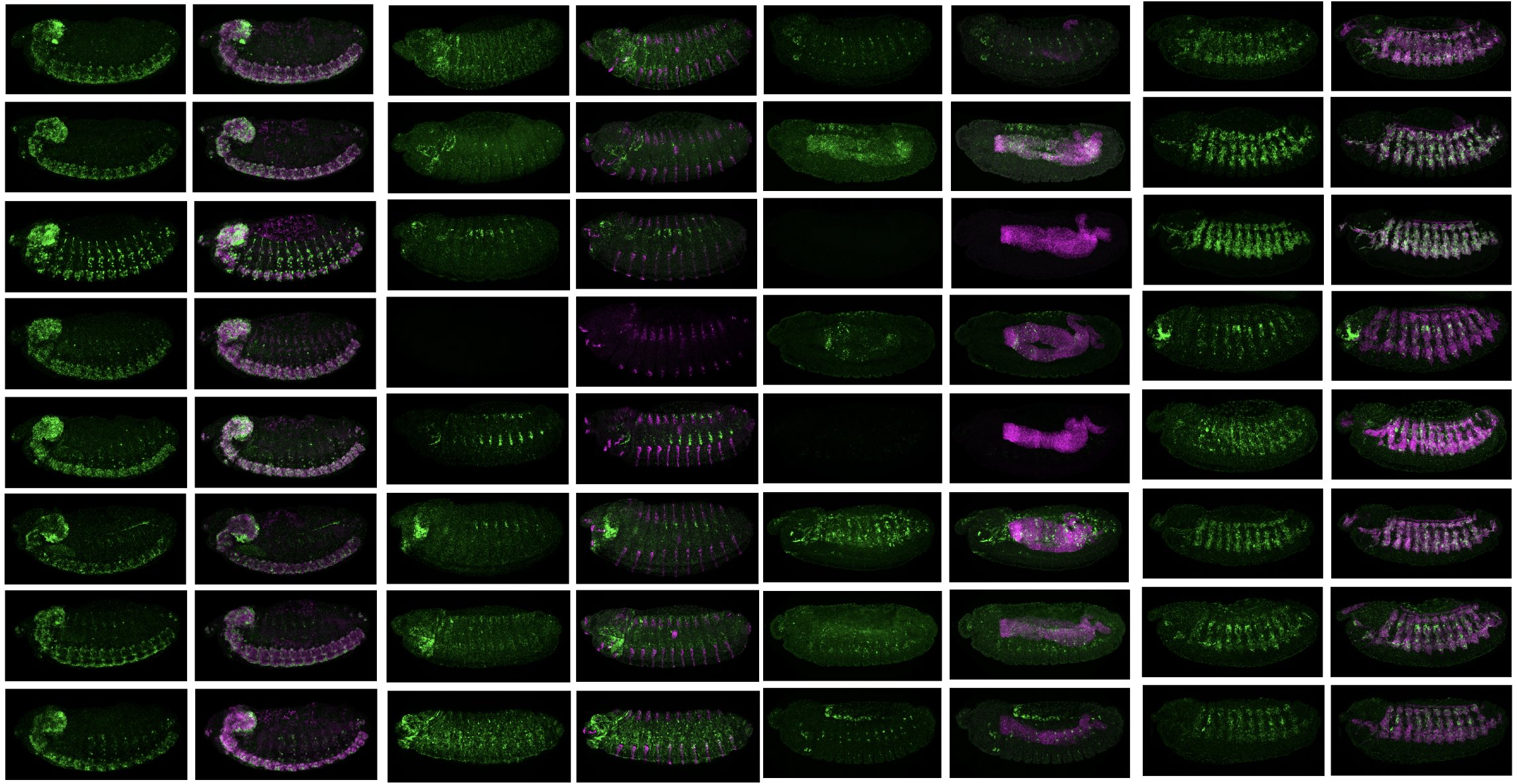
20. F. Reiter*, B.P. de Almeida*, A. Stark. Enhancers display constrained sequence flexibility and context-specific modulation of motif function. Genome Research 2023
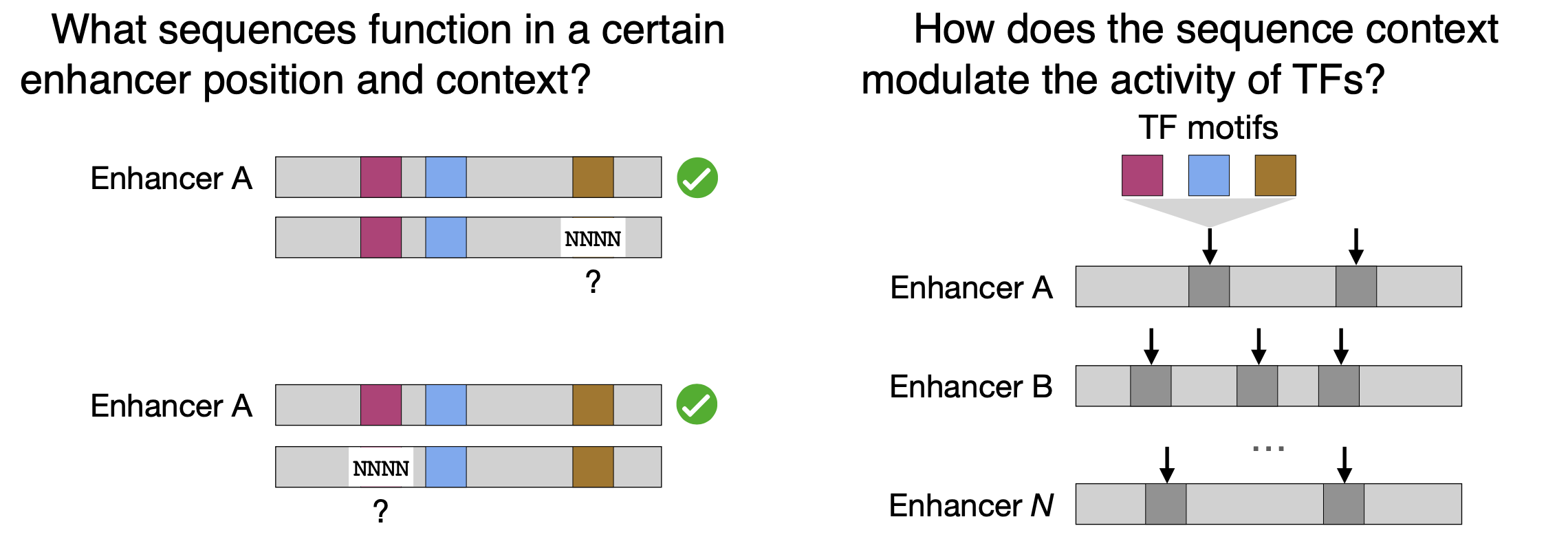
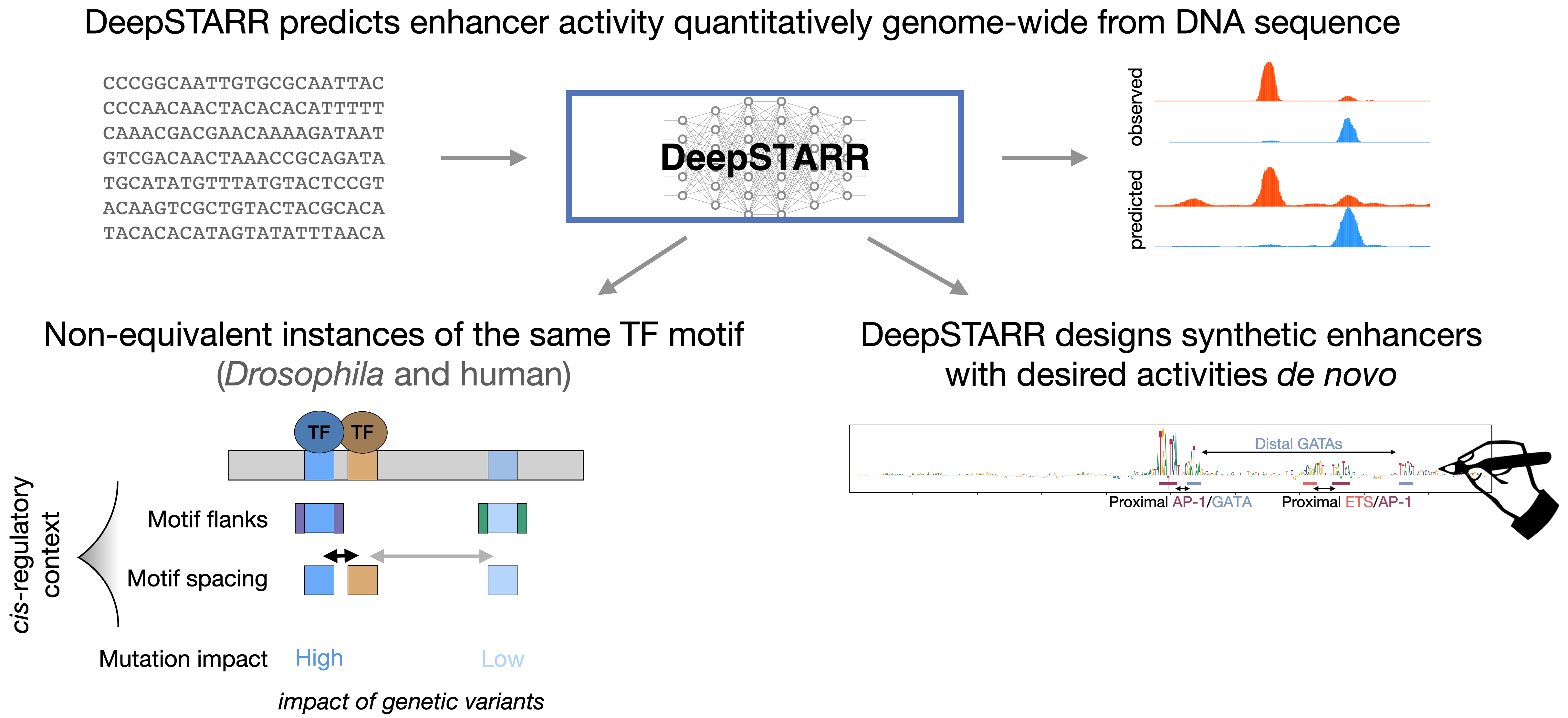
19. B.P. de Almeida, F. Reiter, M. Pagani, A. Stark. DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers. Nature Genetics 2022
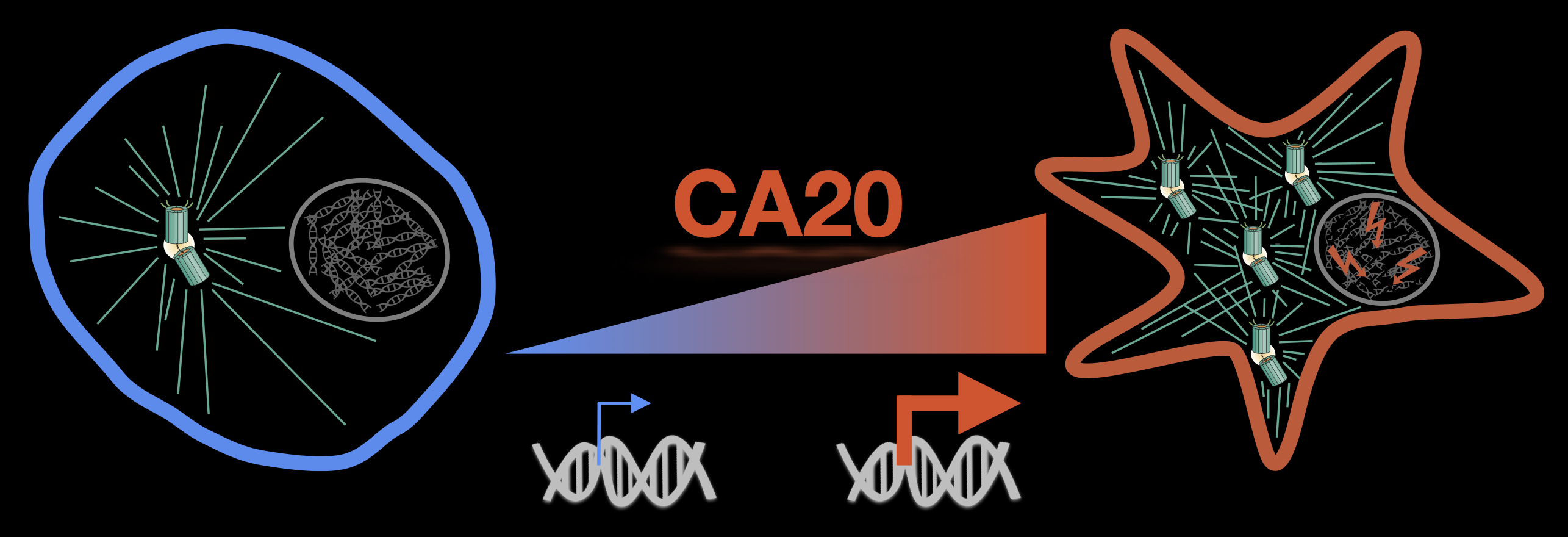
18. B.P. de Almeida, A.F. Vieira, J. Paredes, M. Bettencourt-Dias, N.L. Barbosa-Morais. Pan-cancer association of a centrosome amplification gene expression signature with genomic alterations and clinical outcome. PLoS Computational Biology 2019
17. B.P. de Almeida*, J.D. Apolonio*, A. Binnie, P. Castelo-Branco. Roadmap of DNA methylation in breast cancer identifies novel prognostic biomarkers. BMC Cancer 2019
Preprints
16. B.P. de Almeida*, H. Dalla-Torre*, G. Richard, C. Blum, L. Hexemer, M. Gélard, P. Pandey, S. Laurent, A. Laterre, M. Lang, U. Şahin, K. Beguir, T. Pierrot. SegmentNT: annotating the genome at single-nucleotide resolution with DNA foundation models. bioRxiv 2024

15. J. Mendoza-Revilla, E. Trop, L. Gonzalez, M. Roller, H. Dalla-Torre, B.P. de Almeida, G. Richard, J. Caton, N.L. Carranza, M. Skwark, A. Laterre,K. Beguir, T. Pierrot, M. Lopez. A Foundational Large Language Model for Edible Plant Genomes. bioRxiv 2023
14. H. Dalla-Torre, L. Gonzalez, J. Mendoza-Revilla, N.L. Carranza, A.H. Grzywaczewski, F. Oteri, C. Dallago, E. Trop, B.P. de Almeida, H. Sirelkhatim, G. Richard, M. Skwark, K. Beguir, M. Lopez, T. Pierrot. The Nucleotide Transformer: Building and Evaluating Robust Foundation Models for Human Genomics. bioRxiv 2023
13. V. Loubiere, B.P. de Almeida, M. Pagani, A. Stark. Developmental and housekeeping transcriptional programs display distinct modes of enhancer-enhancer cooperativity in Drosophila. bioRxiv 2023
12. N. Moreno-Marin, G. Marteil, N.C. Fresmann, B.P. de Almeida, K. Dores, R. Fragoso, J. Cardoso, J.B. Pereira-Leal, J.T. Barata, S. Godinho, N.L. Barbosa-Morais, M. Bettencourt-Dias. High prevalence and dependence of centrosome clustering in mesenchymal tumors and leukemia. bioRxiv 2023
11. J.M. Xavier, R. Magno, R. Russell, B.P. de Almeida, A. Jacinta-Fernandes, A. Duarte, M. Dunning, S. Samarajiwa, M. O’Reilly, C.L. Rocha, N. Rosli, B.A.J. Ponder, A.T. Maia. Mapping of cis-regulatory variants by differential allelic expression analysis identifies candidate risk variants and target genes of 27 breast cancer risk loci. medRxiv 2022
Others
10. L. Klaus, B.P. de Almeida, A. Vlasova, F. Nemčko, A. Schleiffer, K. Bergauer, L. Hofbauer, M. Rath, A. Stark. Identification and characterization of repressive domains in Drosophila transcription factors. The EMBO Journal 2022
9. L. Correia, J.M. Xavier, R. Magno, B.P. de Almeida, F. Esteves, I. Duarte, M. Eldridge, C. Sun, A. Bosma, L. Mittempergher, A. Marreiros, R. Bernards, C. Caldas, S.-F. Chin§ and A.-T. Maia§. Allelic expression imbalance of PIK3CA mutations is frequent in breast cancer and prognostically significant. npj Breast Cancer 2022
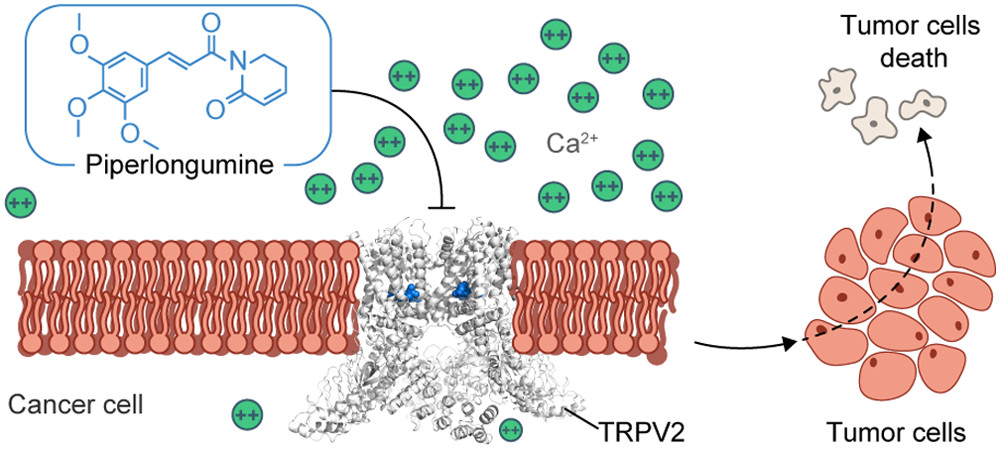
8. J. Conde*, R.A. Pumroy*, C. Baker*, T. Rodrigues*, A. Guerreiro, B.B. Sousa, M.C. Marques, B.P. de Almeida, S. Lee, E.P. Leites, D. Picard, A. Samanta, S.H. Vaz, F. Sieglitz, M. Langini, M. Remke, R. Roque, T. Weiss, M. Weller, Y. Liu, S. Han, F. Corzana, V.A. Morais, C.C. Faria, T. Carvalho, P. Filippakopoulos, B. Snijder, N.L. Barbosa-Morais, V.Y. Moiseenkova-Bell§, and G.J.L. Bernardes§. Allosteric Antagonist Modulation of TRPV2 by Piperlongumine Impairs Glioblastoma Progression. ACS Cent. Sci. 2021

7. I. Gomes, B.P. de Almeida, S. Dâmaso, A. Mansinho, I. Correia, S. Henriques, R. Cruz-Duarte, G. Vilhais, P. Félix, P. Alves, P. Corredeira, N.L. Barbosa-Morais, L. Costa, S. Casimiro. Expression of receptor activator of NFkB (RANK) drives stemness and resistance to therapy in ER+HER2- breast cancer. Oncotarget 2020
6. T. Rodrigues, B.P. de Almeida, N.L. Barbosa-Morais, G.J.L. Bernardes. Dissecting celastrol with machine learning to unveil dark pharmacology. Chemical Communications 2019
5. C. Baker, T. Rodrigues, B.P. de Almeida, N.L. Barbosa-Morais, G.J.L. Bernardes. Natural product– drug conjugates for modulation of TRPV1-expressing tumors. Bioorganic & Medicinal Chemistry 2019
4. S. Braun*, M. Enculescu*, S.T. Setty*, M. Cortés-López, B.P. de Almeida, F.X.R. Sutandy, L. Schulz, A. Busch, M. Seiler, S. Ebersberger, N.L. Barbosa-Morais, S. Legewie, J. König, K. Zarnack. Decoding a cancer-relevant splicing decision in the RON proto-oncogene using high-throughput mutagenesis. Nature Communications 2018
3. G. Marteil, A. Guerrero, A.F. Vieira, B.P. de Almeida, P. Machado, S. Mendonça, M. Mesquita, B. Villarreal, I. Fonseca, M.E. Francia, K. Dores, N.P. Martins, S.S. Jana, E. Tranfield, N.L. Barbosa- Morais, J. Paredes, D. Pellman, S.A. Godinho, M. Bettencourt-Dias. Over-elongation of centrioles in cancer promotes centriole amplification and chromosome missegregation. Nature Communications 2018
Workshop papers
2. E. Trop, C.-H. Kao, M. Polen, Y. Schiff, B.P. de Almeida, A. Gokaslan, T. Pierrot, V. Kuleshov. Advancing DNA language models: The genomics long-range benchmark. AAAI, LLMs4Bio Workshop 2024
1. S. Boshar , E. Trop, B.P. de Almeida, T. Pierrot. Exploring Genomic Language Models on Protein Downstream Tasks. AAAI, LLMs4Bio Workshop 2024
PhD Thesis
B.P. de Almeida. Decoding the cis-regulatory information of enhancer sequences. 2023
For an automatically updated version, check Google Scholar, PubMed or ORCID.
Softwares
B.P. de Almeida*, N. Saraiva-Agostinho*, N.L. Barbosa-Morais. cTRAP: Identification of candidate causal perturbations from differential gene expression data. R package
* equal contribution, § shared corresponding author