Publications
Selected


25.
Targeted design of synthetic enhancers for selected tissues in the Drosophila embryo
Featured commentary: Alexandra Despang. "Designer enhancers for cell-type-specific gene regulation". Nature Biotechnology 2024
Artificial Intelligence cracks code of gene regulation
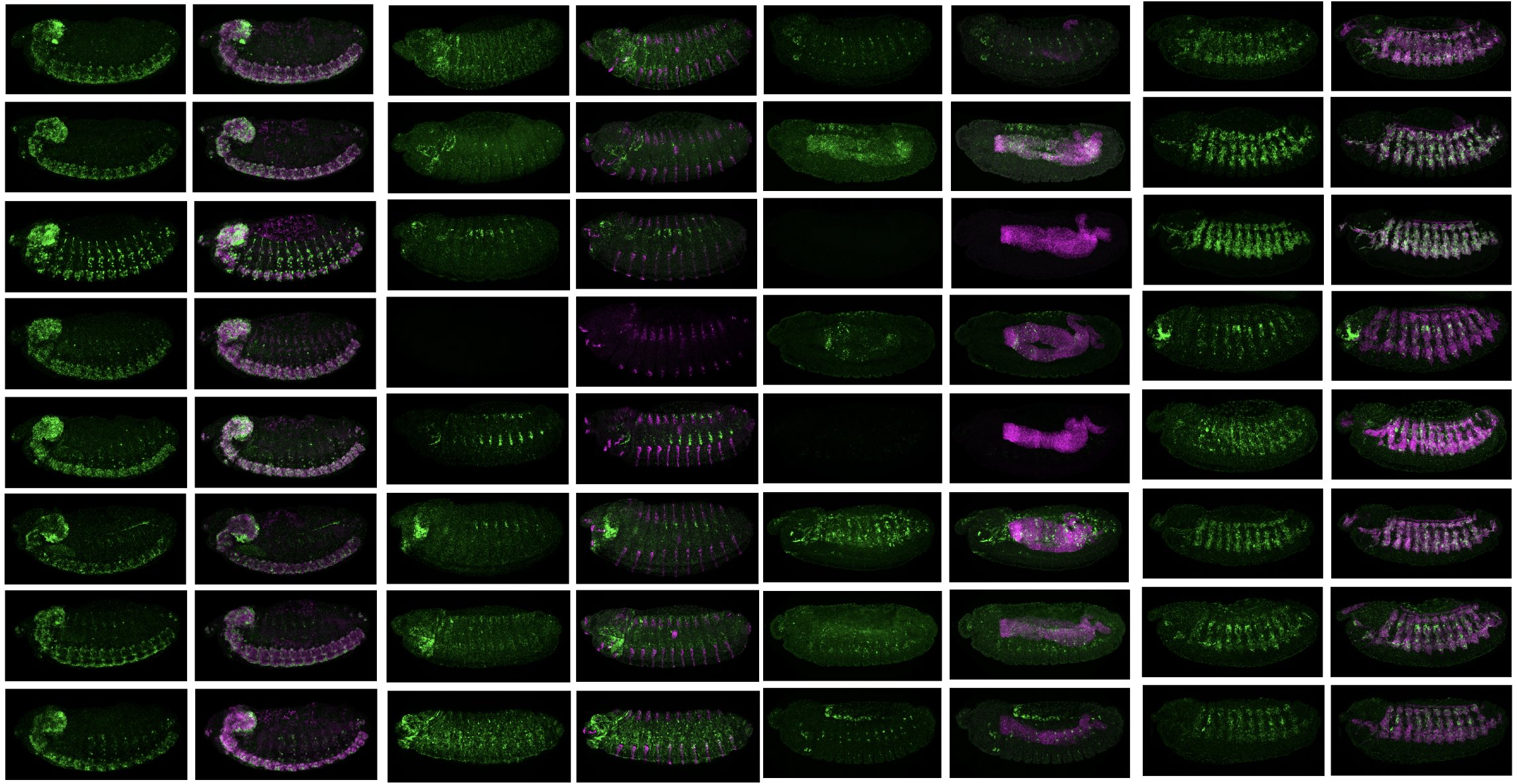
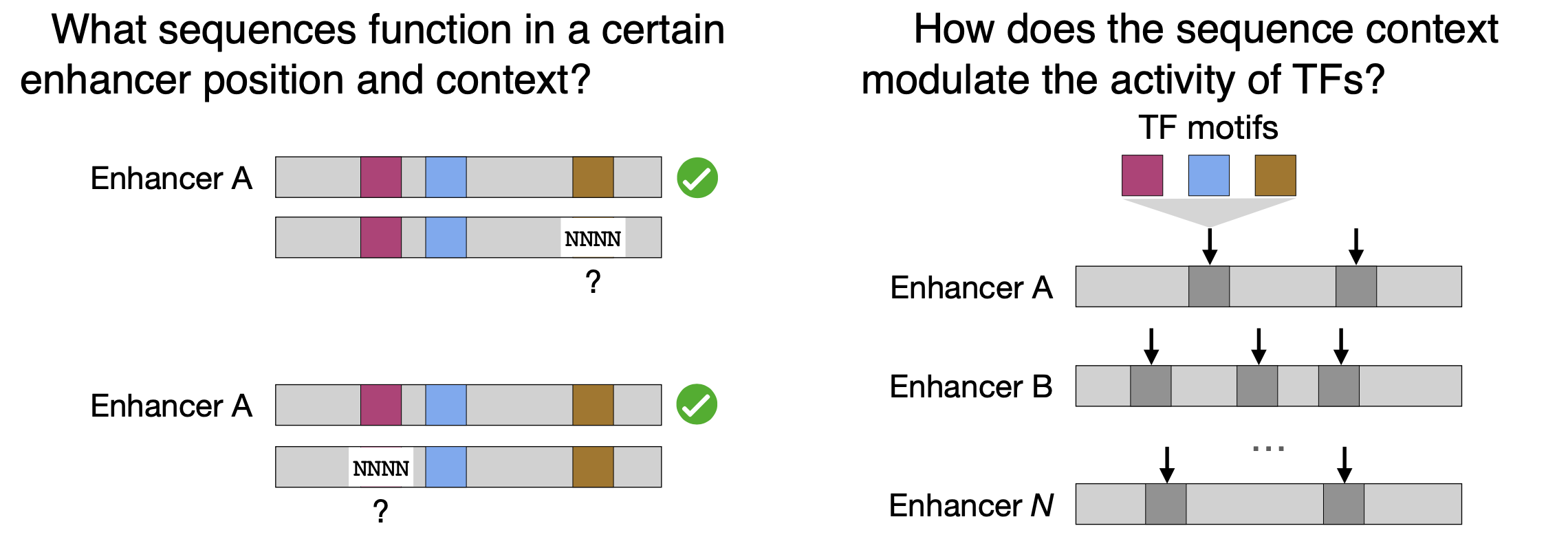
24.
Enhancers display constrained sequence flexibility and context-specific modulation of motif function
Decoding the grammar of gene regulation
Poster at EMBL Conference: Transcription and Chromatin (August 2022)
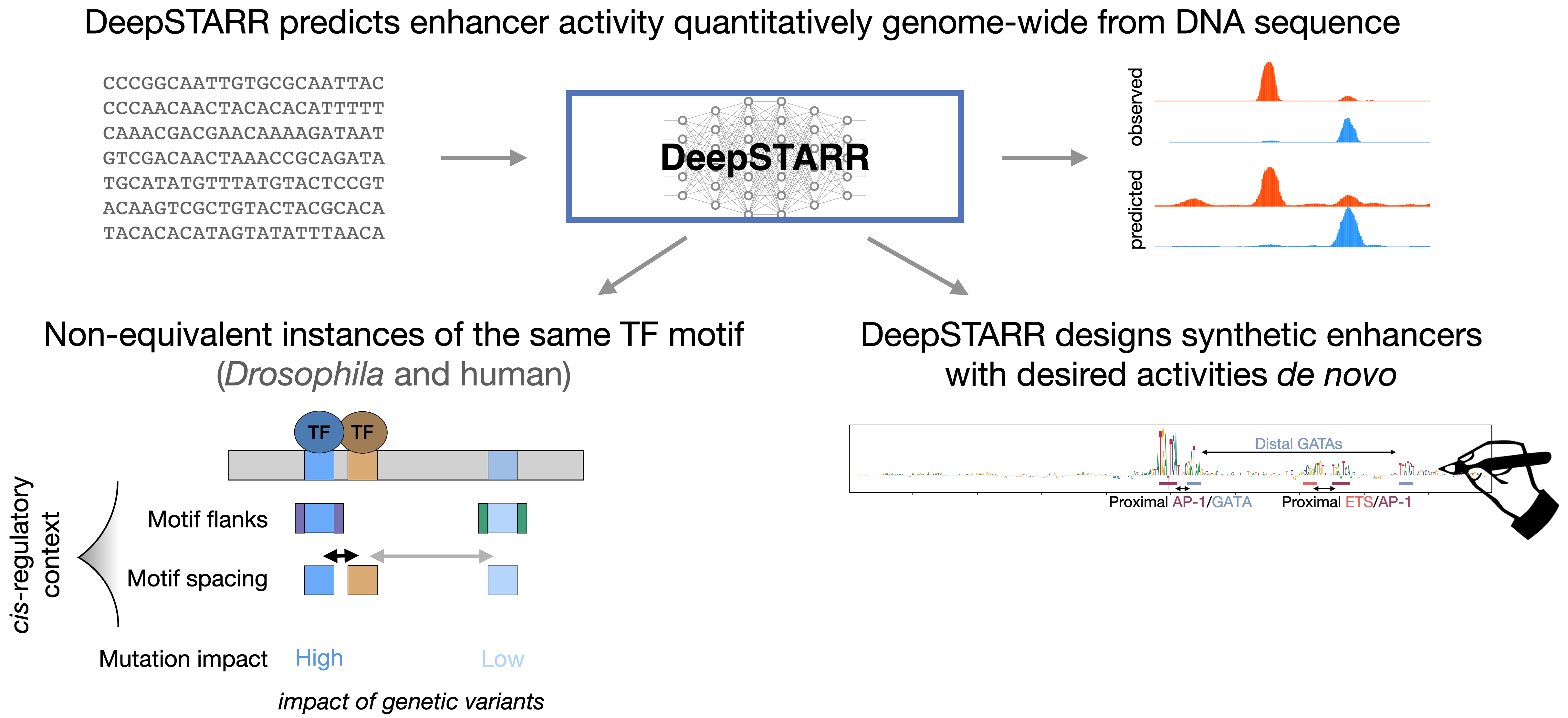
23.
DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers
Featured commentary: Lin Tang. "Predicting and designing enhancers". Nature Methods 2022
Awarded the Life Science Research Award Austria 2022, by ÖGMBT
Harnessing artificial intelligence to predict and control gene regulation
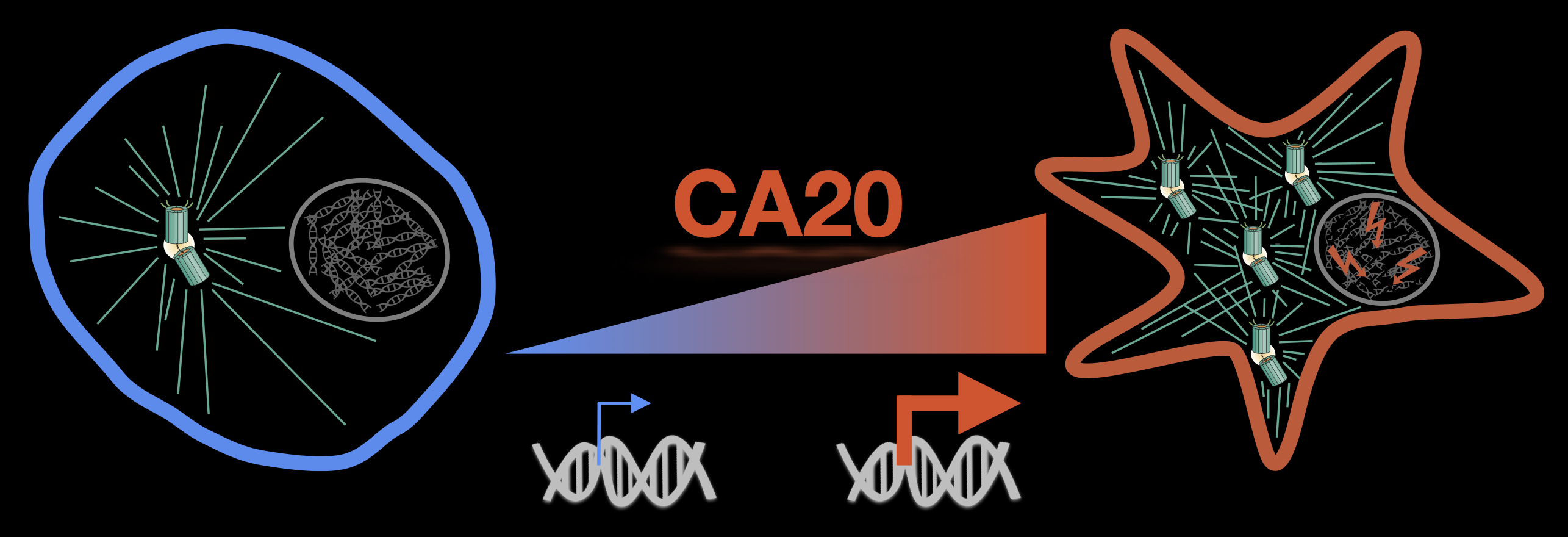
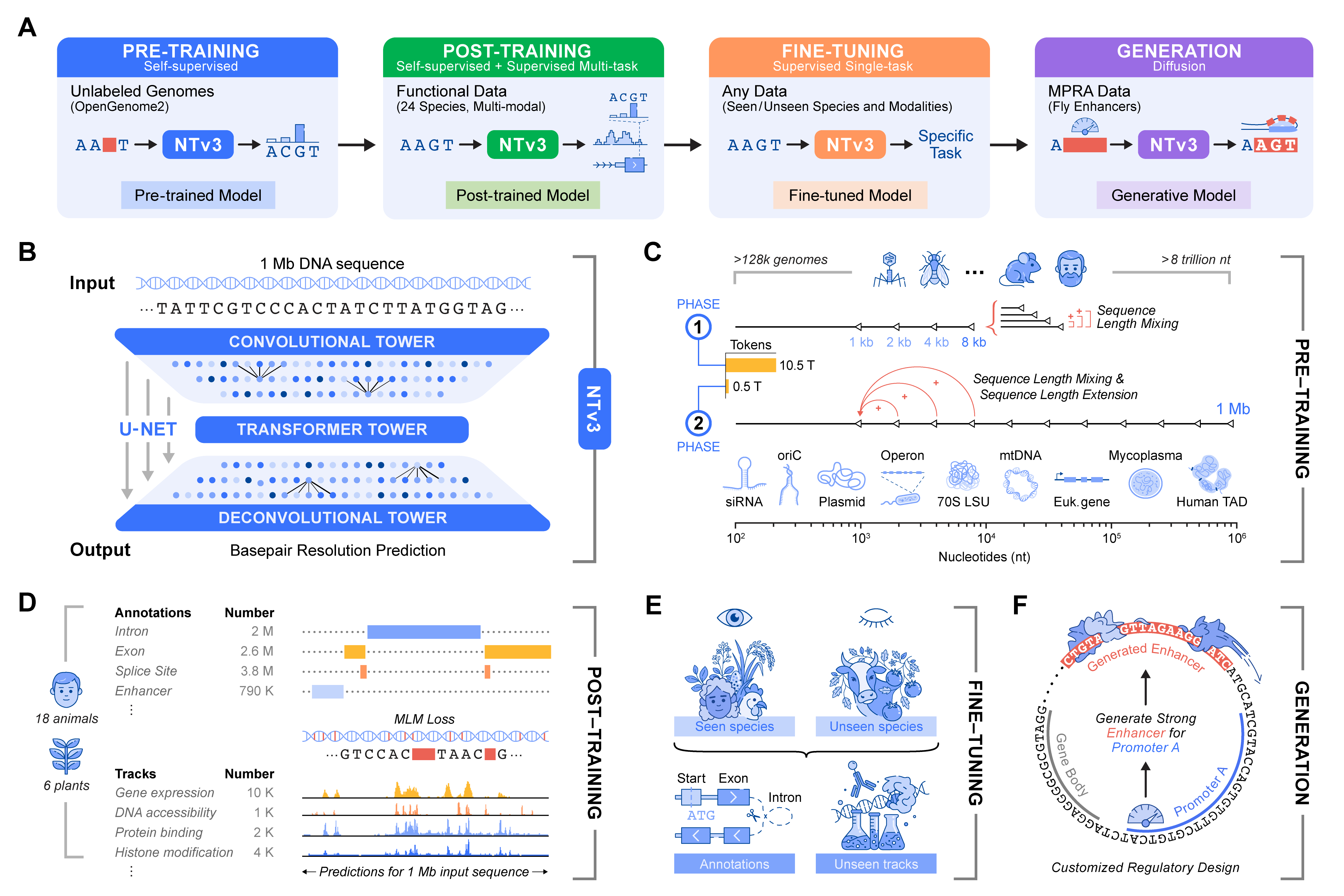
Preprints
Others
Workshop papers
PhD Thesis
Softwares
Patents
* equal contribution, § shared corresponding author